class: center, middle, inverse, title-slide # DSBA 5122: Visual Analytics ## Class 7: Multidimensional & Dimensionality Reduction ### Ryan Wesslen ### October 14, 2019 --- class: center, middle, inverse # Multidimensional Data: Wilke Ch. 12 <img src="../images/slides/07-class/table_lens.png" width="600px" style="display: block; margin: auto;" /> Rao and Card, 1994 --- ## Summary information ```r glimpse(mpg) ``` ``` ## Observations: 234 ## Variables: 11 ## $ manufacturer <chr> "audi", "audi", "audi", "audi", "audi", "audi", "au… ## $ model <chr> "a4", "a4", "a4", "a4", "a4", "a4", "a4", "a4 quatt… ## $ displ <dbl> 1.8, 1.8, 2.0, 2.0, 2.8, 2.8, 3.1, 1.8, 1.8, 2.0, 2… ## $ year <int> 1999, 1999, 2008, 2008, 1999, 1999, 2008, 1999, 199… ## $ cyl <int> 4, 4, 4, 4, 6, 6, 6, 4, 4, 4, 4, 6, 6, 6, 6, 6, 6, … ## $ trans <chr> "auto(l5)", "manual(m5)", "manual(m6)", "auto(av)",… ## $ drv <chr> "f", "f", "f", "f", "f", "f", "f", "4", "4", "4", "… ## $ cty <int> 18, 21, 20, 21, 16, 18, 18, 18, 16, 20, 19, 15, 17,… ## $ hwy <int> 29, 29, 31, 30, 26, 26, 27, 26, 25, 28, 27, 25, 25,… ## $ fl <chr> "p", "p", "p", "p", "p", "p", "p", "p", "p", "p", "… ## $ class <chr> "compact", "compact", "compact", "compact", "compac… ``` --- ## `skimr` package ```r library(skimr) skim(mpg) ``` ``` ## Skim summary statistics ## n obs: 234 ## n variables: 11 ## ## ── Variable type:character ───────────────────────────────────────────────────────────── ## variable missing complete n min max empty n_unique ## class 0 234 234 3 10 0 7 ## drv 0 234 234 1 1 0 3 ## fl 0 234 234 1 1 0 5 ## manufacturer 0 234 234 4 10 0 15 ## model 0 234 234 2 22 0 38 ## trans 0 234 234 8 10 0 10 ## ## ── Variable type:integer ─────────────────────────────────────────────────────────────── ## variable missing complete n mean sd p0 p25 p50 p75 p100 ## cty 0 234 234 16.86 4.26 9 14 17 19 35 ## cyl 0 234 234 5.89 1.61 4 4 6 8 8 ## hwy 0 234 234 23.44 5.95 12 18 24 27 44 ## year 0 234 234 2003.5 4.51 1999 1999 2003.5 2008 2008 ## hist ## ▅▇▇▇▁▁▁▁ ## ▇▁▁▇▁▁▁▇ ## ▃▇▃▇▅▁▁▁ ## ▇▁▁▁▁▁▁▇ ## ## ── Variable type:numeric ─────────────────────────────────────────────────────────────── ## variable missing complete n mean sd p0 p25 p50 p75 p100 hist ## displ 0 234 234 3.47 1.29 1.6 2.4 3.3 4.6 7 ▇▇▅▅▅▃▂▁ ``` --- ## Table based <div id="htmlwidget-0c2fcf8acf203dad1086" style="width:100%;height:302.4px;" class="widgetframe html-widget"></div> <script type="application/json" data-for="htmlwidget-0c2fcf8acf203dad1086">{"x":{"url":"07-class_files/figure-html//widgets/widget_unnamed-chunk-4.html","options":{"xdomain":"*","allowfullscreen":false,"lazyload":false}},"evals":[],"jsHooks":[]}</script> --- ```r library(formattable) head(df) ``` ``` ## id name age grade test1_score test2_score final_score registered ## 1 1 Bob 28 C 8.9 9.1 9.0 TRUE ## 2 2 Ashley 27 A 9.5 9.1 9.3 FALSE ## 3 3 James 30 A 9.6 9.2 9.4 TRUE ## 4 4 David 28 C 8.9 9.1 9.0 FALSE ## 5 5 Jenny 29 B 9.1 8.9 9.0 TRUE ## 6 6 Hans 29 B 9.3 8.5 8.9 TRUE ``` ```r f <- formattable(df, list( age = color_tile("white", "orange"), grade = formatter("span", style = x ~ ifelse(x == "A", style(color = "green", font.weight = "bold"), NA)), area(col = c(test1_score, test2_score)) ~ normalize_bar("pink", 0.2), final_score = formatter("span", style = x ~ style(color = ifelse(rank(-x) <= 3, "green", "gray")), x ~ sprintf("%.2f (rank: %02d)", x, rank(-x))), registered = formatter("span", style = x ~ style(color = ifelse(x, "green", "red")), x ~ icontext(ifelse(x, "ok", "remove"), ifelse(x, "Yes", "No"))) )) # to make work in Rmarkdown/xaringan f %>% as.htmlwidget() %>% frameWidget() ``` --- ## `datacomb` package <img src="../images/slides/07-class/dc2-demo.gif" width="700px" style="display: block; margin: auto;" /> <https://github.com/cmpolis/datacomb> --- ## Heatmaps ```r library(d3heatmap) d3heatmap(mtcars, scale = "column", colors = "YlOrRd") ``` <div id="htmlwidget-283eb981192694f3eb85" style="width:100%;height:302.4px;" class="widgetframe html-widget"></div> <script type="application/json" data-for="htmlwidget-283eb981192694f3eb85">{"x":{"url":"07-class_files/figure-html//widgets/widget_unnamed-chunk-9.html","options":{"xdomain":"*","allowfullscreen":false,"lazyload":false}},"evals":[],"jsHooks":[]}</script> --- ## Scatterplot matrix <img src="../images/slides/07-class/cairo-scattermatrix.png" width="650px" style="display: block; margin: auto;" /> --- ## Scatterplot matrix <img src="../images/slides/07-class/cairo-scattermatrix2.png" width="570px" style="display: block; margin: auto;" /> --- ## Scatterplot matrix ```r library(pairsD3) pairsD3(iris[,1:4], group=iris[,5]) ``` <div id="htmlwidget-d2bc67ea1ffc7f1fda67" style="width:100%;height:302.4px;" class="widgetframe html-widget"></div> <script type="application/json" data-for="htmlwidget-d2bc67ea1ffc7f1fda67">{"x":{"url":"07-class_files/figure-html//widgets/widget_unnamed-chunk-13.html","options":{"xdomain":"*","allowfullscreen":false,"lazyload":false}},"evals":[],"jsHooks":[]}</script> --- ## Parallel coordinates: <img src="../images/slides/07-class/parallel.png" width="620px" style="display: block; margin: auto;" /> <https://dsba5122.com/images/slides/07-class/parcoords.html> [`parcoords`](http://www.buildingwidgets.com/blog/2015/1/30/week-04-interactive-parallel-coordinates-1) --- ## Radar (Star) Plot <img src="../images/slides/07-class/star.png" width="500px" style="display: block; margin: auto;" /> --- ## Radar (Star) Plot <img src="../images/slides/07-class/star2.png" width="600px" style="display: block; margin: auto;" /> --- ## Chernoff Faces <img src="../images/slides/07-class/chernoff1.png" width="700px" style="display: block; margin: auto;" /> --- ## Chernoff Faces <img src="../images/slides/07-class/chernoff2.png" width="700px" style="display: block; margin: auto;" /> --- ## Chernoff Faces: `ggChernoff` ```r #devtools::install_github('Selbosh/ggChernoff') library(ggChernoff) ggplot(data.frame(x = rnorm(20), y = rexp(20), z = runif(20))) + aes(x, y, smile = z) + geom_chernoff(fill = 'steelblue1') + scale_smile_continuous('Smilez', breaks = 0:10/10, midpoint = .5) ``` 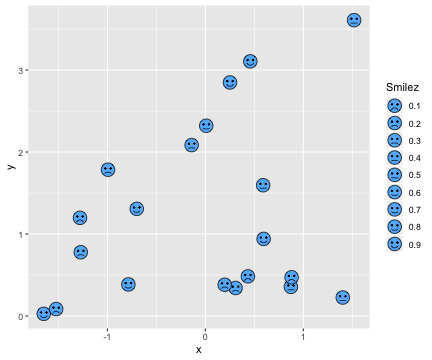<!-- --> --- ## `DfaceR` Shiny App <img src="../images/slides/07-class/faces.png" width="500px" style="display: block; margin: auto;" /> <https://oddhypothesis.shinyapps.io/DFaceR/> --- class: center, middle, inverse # Dimensionality Reduction: <img src="../images/slides/07-class/dim-reduction.png" width="500px" style="display: block; margin: auto;" /> --- # Simplest approach: `dplyr` <img src="../images/slides/07-class/dplyr.png" width="300px" style="display: block; margin: auto;" /> --- ## Dimensionality Reduction There exist many algorithms for projecting n-dimensions to 2D: 1. Principal components analysis (PCA) 2. Multi-dimensional scaling (MDS) 3. Linear discriminant analysis (LDA) 4. t-Distributed Stochastic Neighbor Embedding (t-SNE) 5. Uniform Manifold Approximation and Projection (UMAP) --- ## Principal Components <img src="../images/slides/07-class/pca.png" width="500px" style="display: block; margin: auto;" /> - Introduces a new set of variables (PC's) by **linear combination of the original variables** and standardized (zero mean and unit variance). - The PCs are uncorrelated and ordered (first feature most important, etc.) - Usually, key data features can be seen from first 2-3 PC's. --- ## Case Study 1: Perfect Human <img src="../images/slides/07-class/pca_12.jpg" width="450px" style="display: block; margin: auto;" /> Lior Patcher's [Dec 2014 blog post](https://liorpachter.wordpress.com/tag/principal-component-analysis/) / see [definition of "Repute"](https://www.snpedia.com/index.php/Repute) genes (good/bad) --- ## Case Study 1: Perfect Human <img src="../images/slides/07-class/yuiza_s-lind.jpg" width="450px" style="display: block; margin: auto;" /> Lior Patcher's [Dec 2014 blog post](https://liorpachter.wordpress.com/tag/principal-component-analysis/) --- ## Case Study 2: Tyler Bradley's [blog post](https://tbradley1013.github.io/2018/02/01/pca-in-a-tidy-verse-framework/) ```r us_arrests %>% head() ``` ``` ## # A tibble: 6 x 5 ## state murder assault urbanpop rape ## <chr> <dbl> <int> <int> <dbl> ## 1 Alabama 13.2 236 58 21.2 ## 2 Alaska 10 263 48 44.5 ## 3 Arizona 8.1 294 80 31 ## 4 Arkansas 8.8 190 50 19.5 ## 5 California 9 276 91 40.6 ## 6 Colorado 7.9 204 78 38.7 ``` ```r us_arrests_pca <- us_arrests %>% nest() %>% mutate(pca = map(data, ~ prcomp(.x %>% select(-state), center = TRUE, scale = TRUE)), pca_aug = map2(pca, data, ~augment(.x, data = .y))) us_arrests_pca %>% head() ``` ``` ## # A tibble: 1 x 3 ## data pca pca_aug ## <list> <list> <list> ## 1 <tibble [50 × 5]> <prcomp> <tibble [50 × 10]> ``` --- ## PCA .pull-left[ ```r var_exp <- us_arrests_pca %>% unnest(pca_aug) %>% summarize_at(.vars = vars(contains("PC")), .funs = funs(var)) %>% gather(key = pc, value = variance) %>% mutate(var_exp = variance/sum(variance), cum_var_exp = cumsum(var_exp), pc = str_replace(pc, ".fitted", "")) var_exp ``` ``` ## # A tibble: 4 x 4 ## pc variance var_exp cum_var_exp ## <chr> <dbl> <dbl> <dbl> ## 1 PC1 2.48 0.620 0.620 ## 2 PC2 0.990 0.247 0.868 ## 3 PC3 0.357 0.0891 0.957 ## 4 PC4 0.173 0.0434 1 ``` ] .pull-right[ 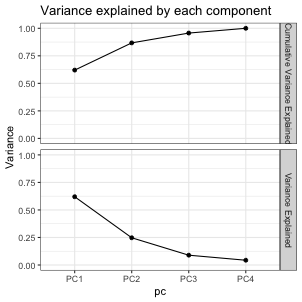<!-- --> ] --- ## PCA with [`ggbiplot`](https://github.com/vqv/ggbiplot) ``` ## [[1]] ``` 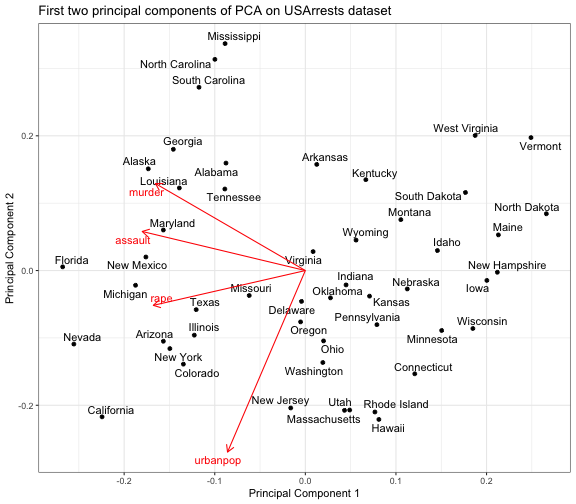<!-- --> --- ## PCA with [`ggbiplot`](https://github.com/vqv/ggbiplot) ```r t <- "First two principal components of PCA on USArrests dataset" us_arrests_pca %>% mutate(pca_graph = map2(.x = pca, .y = data, ~ autoplot(.x, loadings = TRUE, loadings.label = TRUE, loadings.label.repel = TRUE, data = .y, label = TRUE, label.label = "state", label.repel = TRUE) + theme_bw() + labs(x = "Principal Component 1", y = "Principal Component 2", title = t) ) ) %>% pull(pca_graph) ```